Count Summary: all
Christian Mertes
2023-07-07
saveRDS(snakemake, snakemake@log$snakemake)
source(snakemake@params$setup, echo=FALSE)
suppressPackageStartupMessages({
library(cowplot)
})
# opts_chunk$set(fig.width=10, fig.height=8)dataset <- snakemake@wildcards$dataset
workingDir <- snakemake@params$workingDir
fdsLocal <- loadFraserDataSet(dir=workingDir, name=paste0("raw-local-", dataset))
fdsMerge <- loadFraserDataSet(dir=workingDir, name=paste0("raw-", dataset))
has_external <- !(all(is.na(fdsMerge@colData$SPLICE_COUNTS_DIR)) || is.null(fdsMerge@colData$SPLICE_COUNTS_DIR))
if(has_external){
fdsMerge@colData$isExternal <- as.factor(!is.na(fdsMerge@colData$SPLICE_COUNTS_DIR))
}else{
fdsMerge@colData$isExternal <- as.factor(FALSE)
}
devNull <- saveFraserDataSet(fdsMerge,dir=workingDir, name=paste0("raw-", dataset))Number of samples:
Local: 100
External: 0
Number of introns:
Local (before filtering): 1178543
After filtering: 110839
Number of splice sites:
Local: 216897
Expression filtering
The expression filtering step removes introns that are lowly expressed. The requirements for an intron to pass this filter are:
- at least 1 sample has 20 counts (K) for the intron
- at least 5% of the samples need to have a total of at least 10 reads for the splice metric denominator (N) of the intron
plotFilterExpression(fdsMerge) +
labs(title="", x="Mean Intron Expression", y="Introns") +
theme_cowplot(font_size = 16)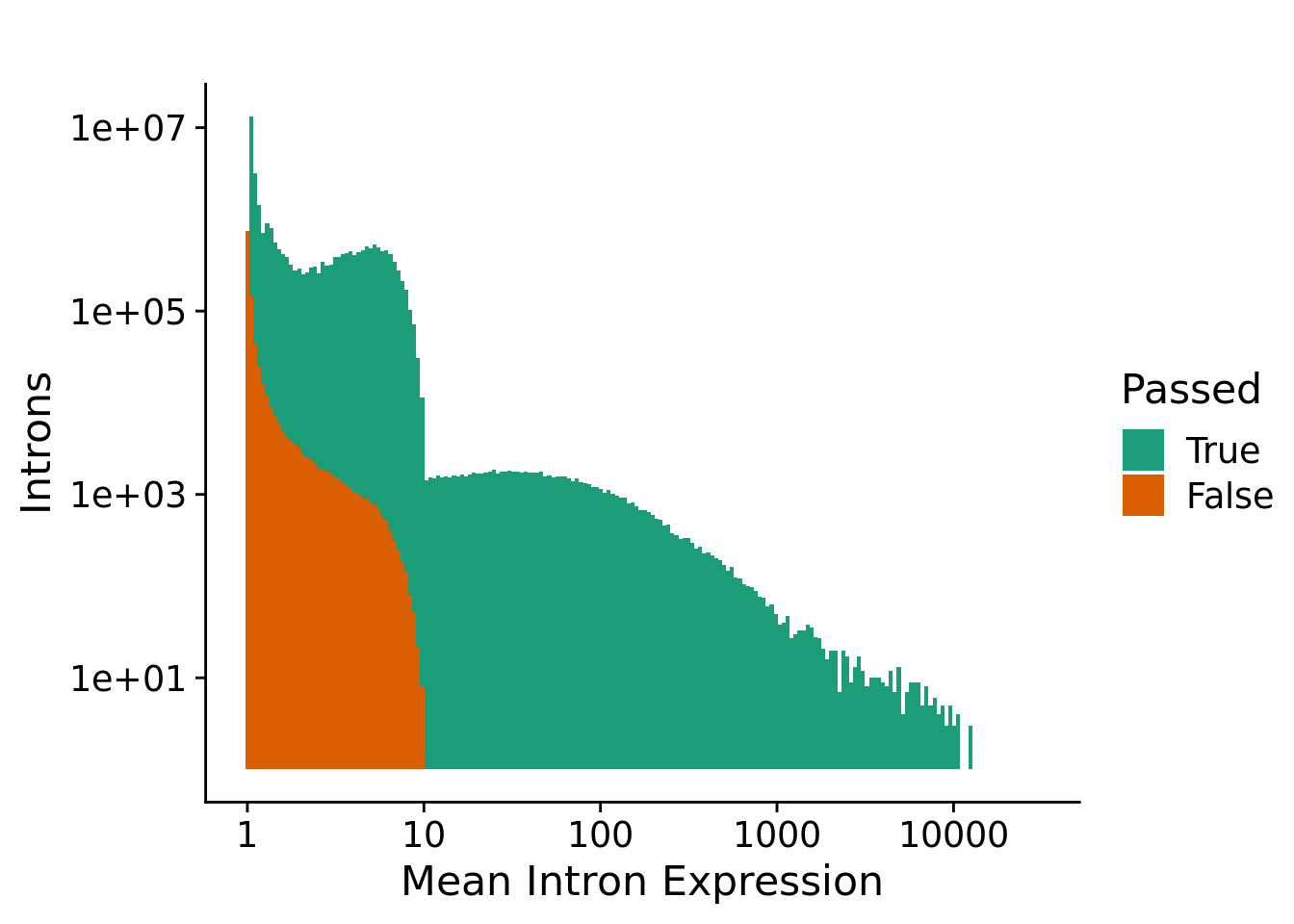
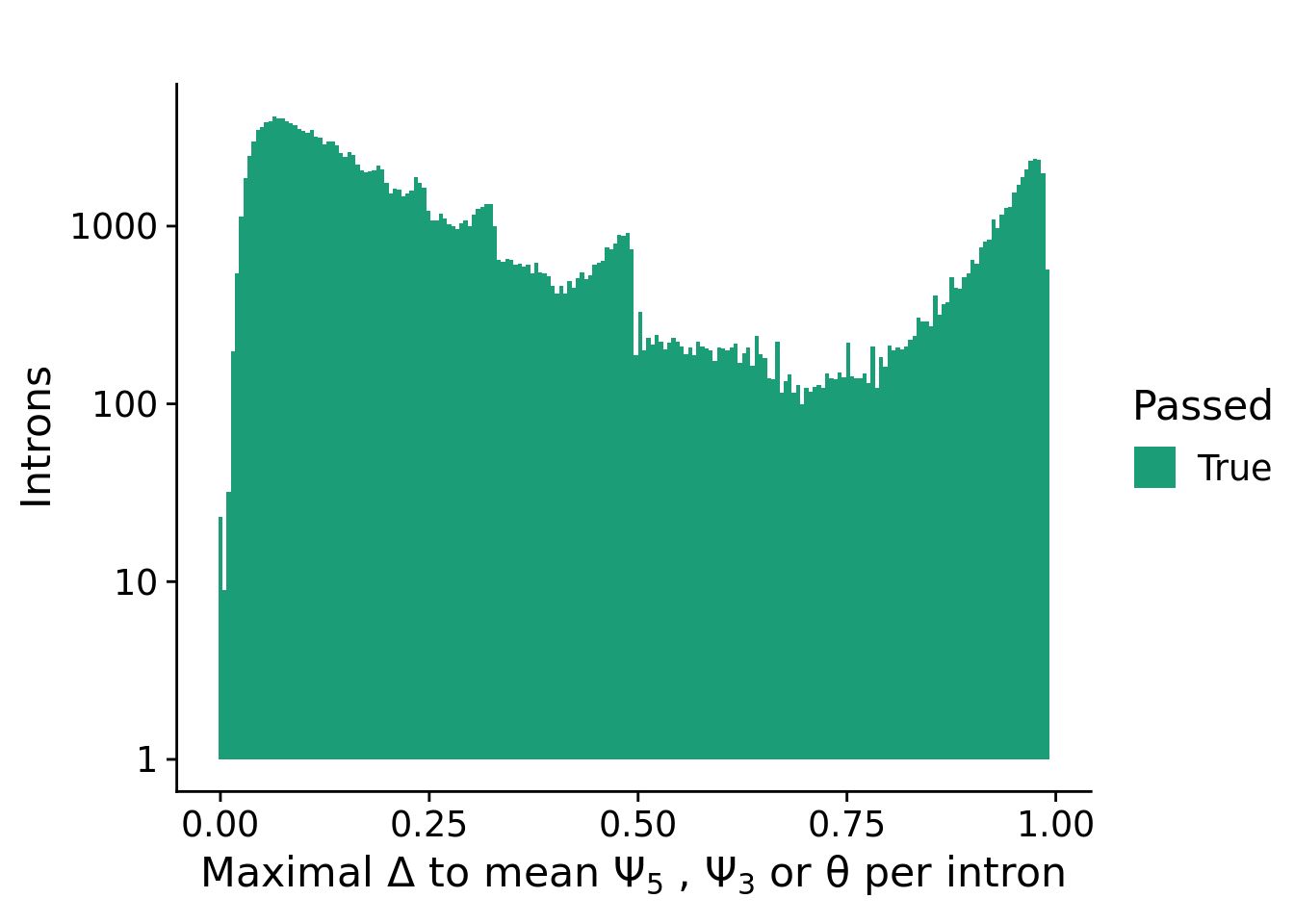
Variability filtering
The variability filtering step removes introns that have no or little variability in the splice metric values across samples. The requirement for an intron to pass this filter is:
- at least 1 sample has a difference of at least 0 in the splice metric compared to the mean splice metric of the intron
plotFilterVariability(fdsMerge) +
labs(title="", y="Introns") +
theme_cowplot(font_size = 16)