Input data and parameters
Input
| Analysis date: | Sun Oct 29 01:47:01 GMT 2023 |
| BAM file: | SRX7354066.markdup.sorted.bam |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | gencode.v44.annotation_genes_collapsed_only_patched_ERCC92.gtf |
| Number of bases for 5'-3' bias computation: | 100 |
| Number of transcripts for 5'-3' bias computation: | 1,000 |
| Paired-end sequencing: | yes |
| Protocol: | strand-specific-reverse |
| Sorting performed: | yes |
Summary
Reads alignment
| Number of mapped reads (left/right): | 40,023,127 / 39,999,985 |
| Number of aligned pairs (without duplicates): | 39,997,496 |
| Total number of alignments: | 87,454,278 |
| Number of secondary alignments: | 7,431,166 |
| Number of non-unique alignments: | 12,993,793 |
| Aligned to genes: | 66,569,187 |
| Ambiguous alignments: | 176,801 |
| No feature assigned: | 7,340,379 |
| Missing chromosome in annotation: | 374,118 |
| Not aligned: | 1,643,388 |
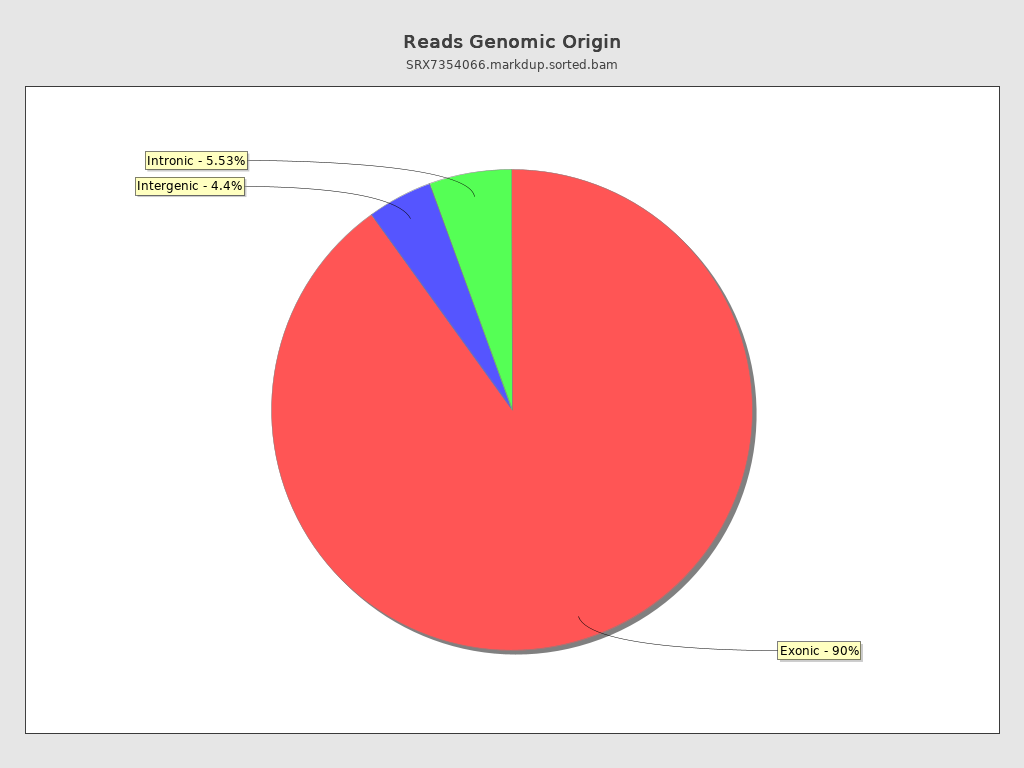
Reads genomic origin
| Exonic: | 66,569,187 / 90.07% |
| Intronic: | 4,089,167 / 5.53% |
| Intergenic: | 3,251,212 / 4.4% |
| Intronic/intergenic overlapping exon: | 2,082,097 / 2.82% |
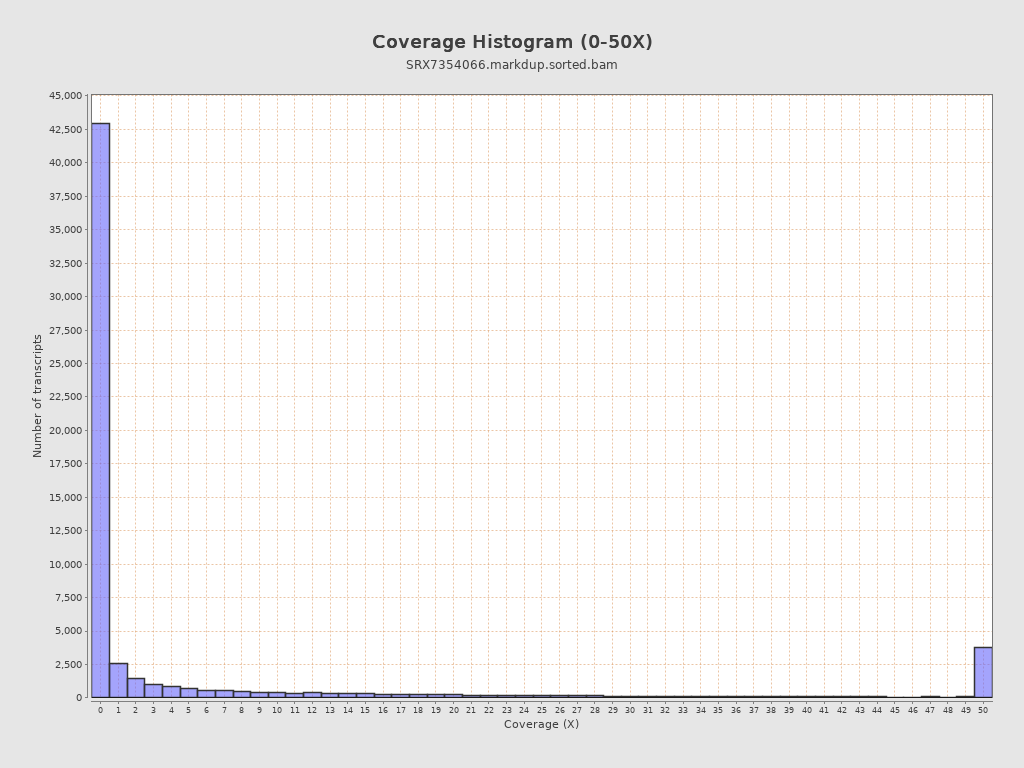
Transcript coverage profile
| 5' bias: | 0.15 |
| 3' bias: | 0.14 |
| 5'-3' bias: | 1.95 |
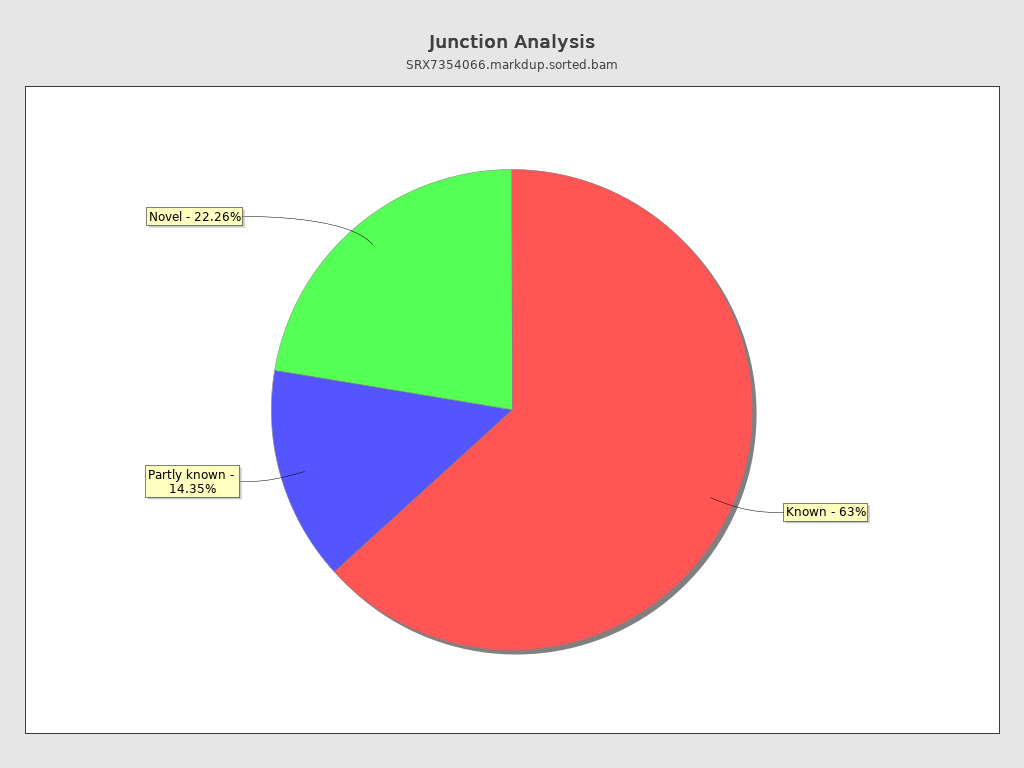
Junction analysis
| Reads at junctions: | 14,228,902 |
| ACCT | 5.14% |
| ATCT | 4.69% |
| AGGT | 4.24% |
| TCCT | 3.8% |
| GCCT | 3.39% |
| AGGA | 3.27% |
| CCCT | 2.91% |
| TTCT | 2.48% |
| AGCT | 2.25% |
| AGGC | 2.12% |
| TCCC | 1.93% |

.png)
.png)
.png)