Input data and parameters
Input
| Analysis date: | Sun Oct 29 12:31:41 GMT 2023 |
| BAM file: | SRX7354053.markdup.sorted.bam |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | gencode.v44.annotation_genes_collapsed_only_patched_ERCC92.gtf |
| Number of bases for 5'-3' bias computation: | 100 |
| Number of transcripts for 5'-3' bias computation: | 1,000 |
| Paired-end sequencing: | yes |
| Protocol: | strand-specific-reverse |
| Sorting performed: | yes |
Summary
Reads alignment
| Number of mapped reads (left/right): | 51,044,898 / 50,992,946 |
| Number of aligned pairs (without duplicates): | 50,983,671 |
| Total number of alignments: | 108,609,243 |
| Number of secondary alignments: | 6,571,399 |
| Number of non-unique alignments: | 10,895,139 |
| Aligned to genes: | 83,032,237 |
| Ambiguous alignments: | 311,458 |
| No feature assigned: | 13,934,617 |
| Missing chromosome in annotation: | 435,792 |
| Not aligned: | 11,688,798 |
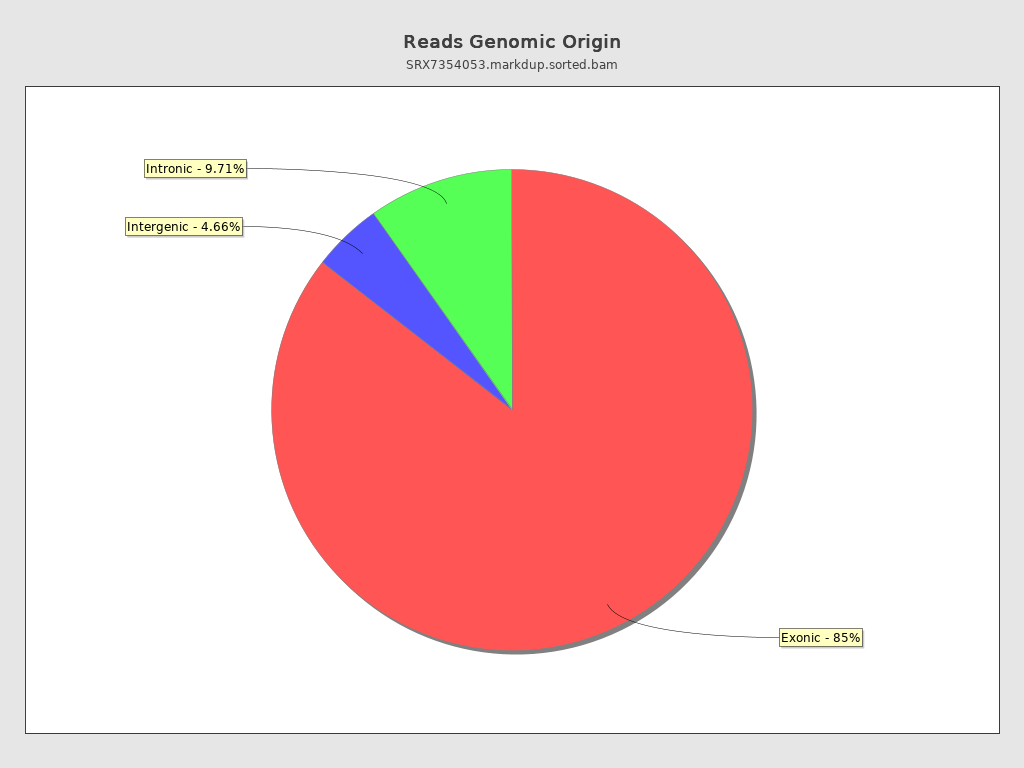
Reads genomic origin
| Exonic: | 83,032,237 / 85.63% |
| Intronic: | 9,417,872 / 9.71% |
| Intergenic: | 4,516,745 / 4.66% |
| Intronic/intergenic overlapping exon: | 3,492,583 / 3.6% |
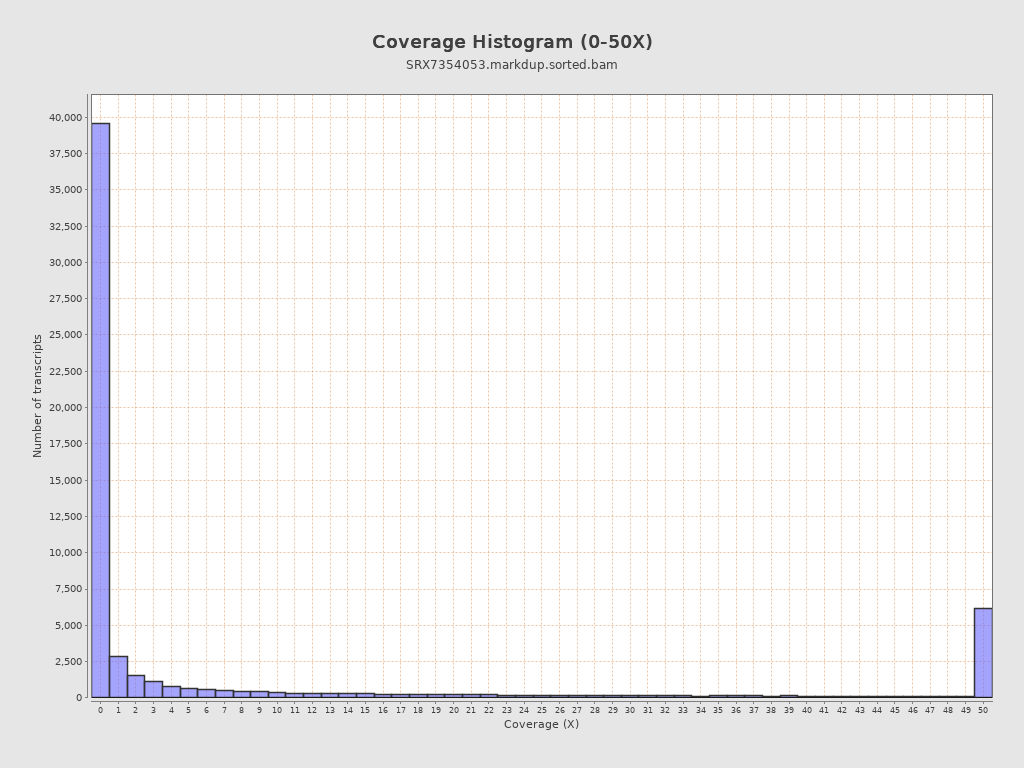
Transcript coverage profile
| 5' bias: | 0.34 |
| 3' bias: | 0.19 |
| 5'-3' bias: | 1.94 |
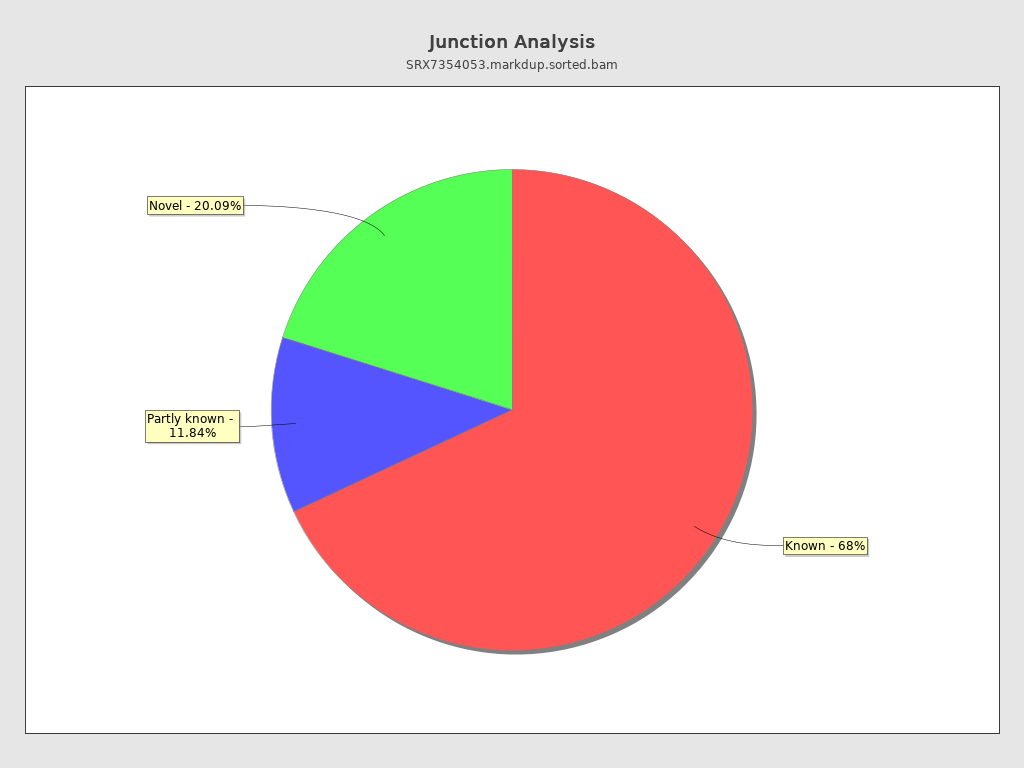
Junction analysis
| Reads at junctions: | 25,645,879 |
| ACCT | 5.32% |
| AGGT | 4.78% |
| AGGA | 3.58% |
| TCCT | 3.56% |
| ATCT | 3.53% |
| GCCT | 2.95% |
| AGCT | 2.65% |
| CCCT | 2.4% |
| AGGC | 2.38% |
| AGAT | 2.31% |
| TTCT | 2.31% |

.png)
.png)
.png)